
- Image via Wikipedia
It’s already quite a while since I’ve started to extensively test performance of HMMER3. As many other people noticed before, speed of the search has improved dramatically – I’m really impressed how fast it is. However, it’s only part of the story. The smaller part actually.
As some of readers may know, most of my projects so far were revolving around protein sequence analysis and sequence-structure relationships. Mainly I was doing analysis of sequences that had no clear similarity to anything known, without functional annotation. Usual task was to run sequence comparison software and look at the end of the hit list, trying to make sense from hits beyond any reasonable E-value thresholds (for example I often run BLAST at E-value of 100 or 1000). I use very limited number of tools, because it takes quite a while to understand on which specific patterns a particular software fails.
The high-end tool I use most often is HHpred – HMM-HMM comparison software. It’s slow but very sensitive – my personal benchmarks show that it is able to identify very subtle patterns in sequence formed slightly above level of similar secondary structures (in other words, from the set of equally dissimilar sequences with identical secondary structure order, it correctly identifies the ones with similar tertiary structure).
The most surprising thing about HMMER3 is that in my personal benchmarks it’s almost as sensitive as HHpred. I wasn’t expecting that HMM-sequence comparison can be as good as HMM-HMM. This observation suggests that there’s still a room for improvement for the latter approach, however it has already big implications.
PFAM will soon migrate to HMMER3 (the PFAM team is now resolving overlaps between families that arose due to increased sensitivity) and the moment it is be available, it will make a huge number of publications obsolete, or simply wrong. There are thousands of articles that discuss in detail evolutionary history of some particular domain (many of these will become obsolete) or draw some conclusions from the observation that some domain is not present in analyzed sequence/system (many of these will need to be revised). It will also make my skills quite obsolete, but that is always to be expected, no matter in what branch of science one is working. I also imagine that systems biology people will be very happy to have much better functional annotation of proteins.
I don’t want to call development of HMMER3 a revolution, but it will definitely have similar impact on biology as BLAST and HMMER2 had. Not only because of its speed, but also because it will create a picture of similarities between all proteins comparable to the picture state-of-the-art methods could only calculate for their small subset.
Related articles by Zemanta
- The curse of BLAST (mndoci.com)
![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_e.png)
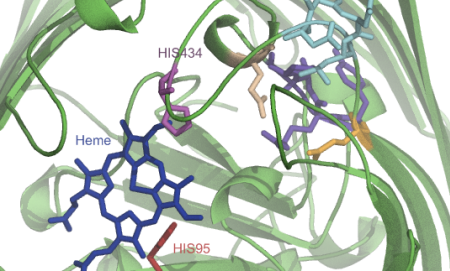
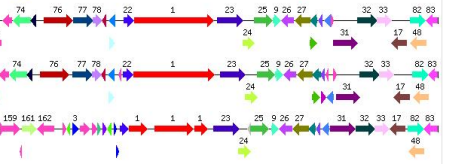
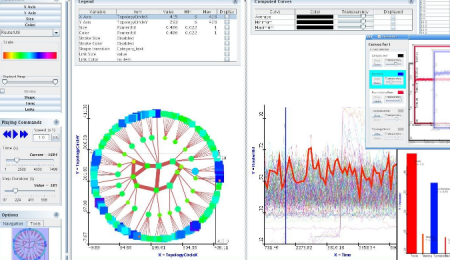
 My recent post on visual analytics in bioinformatics lacked a specific example, but I’m happy to finally provide one (happiness comes also from the fact that respective publication is finally in press). The image above shows a multiple pairwise alignment from BLAST of a putative inner membrane protein from
My recent post on visual analytics in bioinformatics lacked a specific example, but I’m happy to finally provide one (happiness comes also from the fact that respective publication is finally in press). The image above shows a multiple pairwise alignment from BLAST of a putative inner membrane protein from 
















Bug tracking systems in science
I’m not going to describe painful process of correcting entries in biological databases or errors in publications when one is not the author – we all know how difficult and unrewarding it is. All major databases contain wrong entries – I see misannotated (or nonexistent) genes in Genbank, artificial domains in PFAM or poorly solved structures in PDB. It’s even worse in publications, where across the whole spectrum of journals I see errors which in theory shouldn’t slip through peer review (this includes such prominent publishers like NPG).
One of the best idea I heard that addressed this issue was to build a bug tracking system (I would like to give credit to the author, but I cannot find the source; wasn’t that one of biobloggers?). It’s simple and efficient. Something is wrong? Fill a bug report. It would be linking to the original entry, would be available for aggregation (for example to track report’s author activity), and possibly could be closed by somebody else than database maintainers or authors if it’s wrong. Because it would be external to all databases, maybe it could grow to provide “community corrected” versions of these databases?
What do you think? How useful such system could be?
Posted by Pawel Szczesny on April 18, 2008 in Comments, Community, Software
Tags: bioinformatics, bug tracking, NPG, science