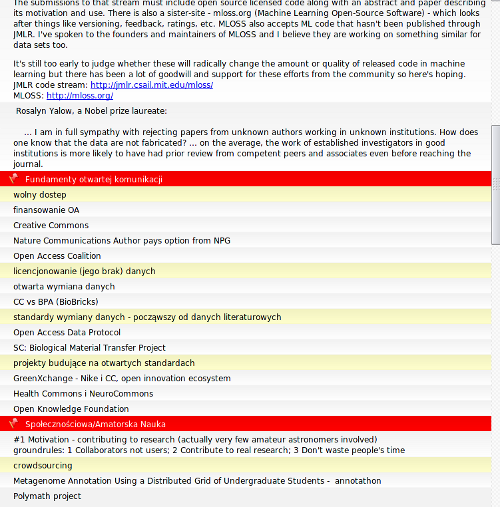
This post is directly inspired by excellent essay by Michael Nielsen entitiled “The Future of Science“. While Michael writes about science itself (and how openness will be playing big role in scientific process) I wanted to write few words about how and where I see scientists in a near future (or rather how the research will be done – I’m not even touching the broad topic of alternative careers for scientists). While it sounds like a complementary essay to Michael’s work, I wouldn’t dare to call it so – think of it as a collection of loose notes gathered over months of learning from online science community. Also, please keep in mind that it’s written by a biologist and as such biased towards life sciences.
It’s no news that academic environment has changed so much that a joy of research spans only small fraction of day-to-day scientists’ life. “Publish or perish“, bureaucracy, money hunting, lack of tenure track positions, impact factor, ever-postdoc are only few of many issues within academic system. There’s quite a lot of interesting initiatives that aim at improving the system and some of them will certainly succeed by solving directly some of the issues above or more likely, by creating a niche within academia in which these issues will not apply. However, I think in the long run academia is not going to be the main environment where the research is being done and more importantly, there will be infinite gradation of research jobs, allowing people from many different fields with many different skills to contribute to scientific projects.
That said, I also believe that amount of data and knowledge produced will lead to enormous specialization of scientists. This does not contradict the previous statement: I don’t think that some teenager will design and develop in his spare time a new molecular dynamics algorithm, but finding new genetic associations or inventing another way to modify bacterial genome so it has better biodegradation features sounds to me like a reachable project for many people. Specialization will be one of many factors influencing creation of new types of scientists. And what are these types? Let me describe a few.
Mind, brain, intelligence amplification – future Nobel Prize winners
This category emerged pretty recently, after reading Deepak’s post on uniqueness (or lack of it) of someone’s contribution to science. I always had this notion that no matter what I did, it would be done in a near future by someone else, but this time I could put it into words: science is like sports – winner takes it all and there’s always a winner. Because prestige of an institution or fame of a scientist plays a big role in getting one’s research funded, competition for money will lead to development of procedures that will aim at producing Nobel Prize winners (or equivalents) analogous to sports training programs.
Techniques like neurolinguistic programming, biofeedback or binaural bits (just to name a few) are surrounded by such a hype, that it’s hard to believe they are worth something. However I think there’s a solid field emerging from these inventions that aims at dealing with issues we create in our lives. Have you heard that Google had opened School of Personal Growth as a part of the Google University, teaching things like mental development, emotional development, holistic health, well-being and finally a Buddhist notion, “beyond the self”? I think it’s no mistake – it’s an attempt to help employees to consistently work at their optimal speed. And there’s story is published by Nature in April last year results of a poll of using brain-doping drugs among scientists. And there’s an inspiring talk by Juan Enriquez on arrival of Homo evolutis. I believe it’s just a matter of time big universities will launch (probably secretly) their own programs for training high profile scientists. And judging from the comments to the Nature’s poll I don’t think many people will object – science, unlike sports, doesn’t have to pretend it’s fair.
Getting research done – staff scientist
This type doesn’t require introduction. If one doesn’t have to waste time on advancing career and hunting for money, one becomes a very efficient scientist. Staff scientist positions are available in many countries and I wish it could be more of them in the future, especially in bioinformatics – where a single person can be trained to do everything from microarrays analysis to molecular dynamics in a relatively (!) short time (and become then a very important asset in the lab).
Experienced specialist – nomadic freelancer
This is category I was aspiring to. Here you can read little details how I tried, and here when and why it failed. I still think it can be done, although not in every field and not all the time. My hope was that telecommuting is the future of freelance scientists, but Bora offered entirely different solution: co-researching spaces/science hostels:
A coworking space has three important components: the physical space, the technological infrastructure, and the people. A Science Hostel that accommodates people who need more than armchairs and wifi, would need to be topical – rooms designed as labs of a particular kind, common equipment that will be used by most people there, all the people being in roughly the same field who use roughly the same tools.
From what I’ve seen, people doing structural biology (especially NMR-related research) tend to enjoy similar to a freelancer status: they can do a crucial high tech task, which takes no more than several weeks to finish and often the task is needed so rarely that there’s no point in employing the specialist full time (or to do in-house training).
The main disadvantage of this mode is something called “consultant’s dilemma” (hat tip Harold Jarche): when you’re working you’re not generating new ideas or business, and vice versa.
In a failure of interdisciplinary approach – translator, integrator
I expect that lots of people will disagree with me on that, but I think on the long run interdisciplinary approaches are going to fail. The area where a reason for failure is most visible is genome sequencing. Deep knowledge about single simple organism such as bacteria is beyond capability of most (if not all) laboratories and teams and that’s why publishing a genome is just a starting point, not end to a process. It takes years of work of experts in their own small fields to extract all useful information from the single sequence.
Once this situation becomes more of an issue, scientific translators may emerge. Such person will track scientific literature in two (or three or four, such as language translators) small fields and will tell group of researchers from one field what important has been published in other field. Will similar service become part of libraries or such people will become independent consultants? I have no idea.
I don’t think that gaps in knowledge will be corrected by talking to colleagues or by review process. Here’s a perfect example (in used-to-be prestigious journal): neither authors nor reviewers have noticed that the structure containing so-called trimerization “octads” is a perfectly fine, quite regular, heptad-based coiled-coil (you guess it right, these “octads” were separated by six residues, giving together fourteen – two coiled-coil heptads). It was already visible in the sequence figure – but only if you knew that things like coiled-coils exist and were already studied by Francis Crick. After almost a year and a half correction wasn’t submitted which means the community does not care either.
Bioentrepreneur
As soon as we have our own Paul Graham and a clear, well-described path of how to make a startup in life sciences successful, we will have a bloom of bioentrepreneurs. Life science is a field comparable to high-tech, not software industry. It requires different skills and different approach, but no one has so far put it into words that we can follow. Also, we need more hardware providers in area of life sciences. If you want to build a mobile phone, it’s a matter of days to order its every single part. If you want to build your own sequencing machine, I wish you good luck, because it will take considerably longer (you need to wait until respective companies are built and offer their products).
Nevertheless, I’m sure it will happen. Streamlining life sciences is something that lots of people are talking about.
Clean data needed – biocurator
The more data the more errors. Recently, I’ve stumbled upon interesting functional annotation of a protein: will die slowly. Search on NCBI reveals few dozens of proteins with such annotation. This is a terse description of a phenotype, however I don’t think should be used as a protein name. Paul Davis suggested that this propagated from Drosophila, since fruit fly gene names have a long history of names blurb:
Early work refers to the gene as fruity, an apparent pun on both the common name of D. melanogaster, the fruit fly, as well as a slang word for homosexual. As social attitudes towards homosexuality changed, fruity came to be regarded as offensive, or at best, not politically correct. Thus, the gene was re-dubbed fruitless, alluding to the lack of offspring produced by flies with the mutation.
It’s nothing new that to reach holy grail of many fields (text mining, ontologies, automated discoveries, predictions), we need manual curation of biological data (even Wolfram Alpha is based on curated data). Similarly to staff scientists, biocurator jobs are already appearing in science job listing.
Science as creative hobby – “not even a scientist”
In the introduction I’ve mentioned a teenager inventing new genetic modification of an organism. While to some it may sound difficult, unquestionable success of iGEM competition shows that it doesn’t require 20 years of research experience to come up with such ideas. Lots of knowledge and lots of data create opportunity for people outside academia to jump in and make a valuable contribution. The necessary requirement in “openness” – as long as the data and publications are freely available, there’s a space for outsiders.
I expect (or I hope) amateur science to grow in the following years – especially in the less bureaucratic countries. If we don’t see many of such examples yet, it’s the education system to blame – kids don’t realize that remixing data and remixing video are very similar things that differ only by a target audience, but both can be cool :).
Knowing your position – “lighthouse” scientist
Lighthouse’s primary role is to assists in navigation – it helps you find your position on the map. Lighthouse is not a point of reference – as a point on the map is usually no more important than any other points. Lighthouse helps you understand where you are. Tech crowd has its own “lighthouse” people, for example Tim O’Reilly. Our small online science community has Bill Hooker. Neither of them seem to have outstanding resume (sorry to write that, I’ve seen better ones), but to understand where you are it’s worth to pay attention to what they say. They seem to understand particular part of our world much better than anybody else.
To put it in other words, a lighthouse scientist isn’t necessarily a person with the biggest achievements or a person who has a brilliant vision of the future – it’s a person who sees trends and movements, has a wider perspective and most importantly knows what’s important. In recent discussions on the blogosphere about bioinformatics as a field of science, Sean Eddy didn’t express his opinion – which I think is a very meaningful response.
Final thoughts
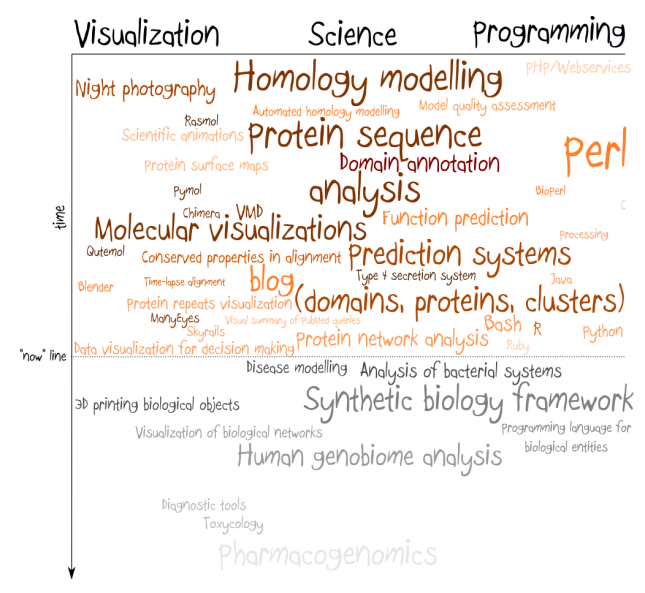
I’ve sketched this map to organize lots of thoughts and discussions around future directions of science. It is far from being complete and full of wishful thinking, but still helped me to wrap my mind around couple of issues in this area. Probably the most important thing I’ve realized is what was put into introduction: that the future may open lots of options for people willing to stay close to science. Those who realize this will benefit from them as first.
Update: there are interesting comments over at FriendFeed already.
 This strategy is similar to the workflow described by Steven Johnson, but without DevonThink. So far I haven’t found anything better under Linux, but probably I need to check online apps – things do change every month.
This strategy is similar to the workflow described by Steven Johnson, but without DevonThink. So far I haven’t found anything better under Linux, but probably I need to check online apps – things do change every month.![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_e.png)



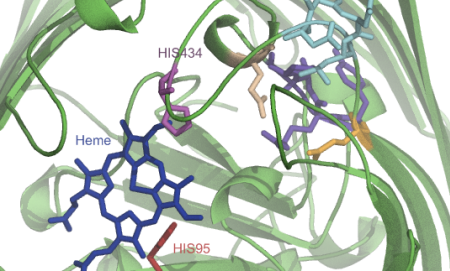
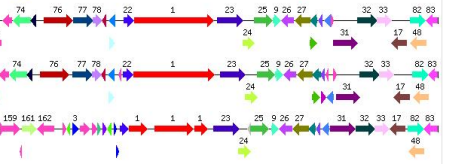
 My recent post on visual analytics in bioinformatics lacked a specific example, but I’m happy to finally provide one (happiness comes also from the fact that respective publication is finally in press). The image above shows a multiple pairwise alignment from BLAST of a putative inner membrane protein from
My recent post on visual analytics in bioinformatics lacked a specific example, but I’m happy to finally provide one (happiness comes also from the fact that respective publication is finally in press). The image above shows a multiple pairwise alignment from BLAST of a putative inner membrane protein from 















Transitions, transitions
Quite a few things happened while I was away. If you’re interested, here’s not so short summary of my internet hiatus:
Research area
I think I’m done with bioinformatics. My current research area seems to be located somewhere between systems biology, theoretical biology and information/complex systems theory. I hope to build on Dawkins work, deal with emergence in biology and study subtle effects in biological systems. While I’m not sure if I will have anything interesting to show ever, I don’t have energy to do yet another project which involves programming/web interfaces/dealing with data/annotations/modelling etc. I’m done with analytics, time for synthesis :).
Carrer
Last year I wrote a post dreaming about small non-profit contract research organisation. This model of Research-as-a-Service has materialized in a virtual research institute which we have finally launched few days ago (materialized in something virtual, sign of times? 😉 ). The setup is quite simple – the institute gets a project (or applies for such) and then it searches for researchers/institutions/freelancers which are willing to subcontract parts of the project. We have outsourced not only research part, even money gathering (writing grants, etc.) is done by external company. The setup is quite flexible and pretty transparent – for example, we may represent somebody’s rights, but no intellectual property is owned by the institute. Why such institution? We become a single point of contact for a large and diverse group of scientists, which are willing to do some research for real money but don’t have time and energy to hunt for gigs by themselves. While I have an academic job, I’m in the middle of transition from being a freelancer, to being a jobs provider for freelance scientists. More on that in some other post.
Open science
I plan to spend way more time on advocating open science (all of its flavors), but… in Polish. This step is out of large frustration that even prominent figures in Polish science have no idea about changes in the science internet-aware researchers are watching and creating. Knowledge about even basic things like Open Access is dramatically low in Poland (a number of people here equals OA with low quality publications which have not been peer-reviewed). With few friends, we have a number of projects in the pipeline (for example, we hope to launch a nation-wide, created by professionals promotional campaign – bilboards, TV commercials etc. – for open science). If any of these actually works, I will let you know if we have any measureable success 😉 .
Labels, labels
Robert Anton Wilson tells a nice story in his book Prometheus Rising:
Another story is a comment from my advisor about putting my real research plans in some proposal (he supports these plans):
I feel like screaming “Labels, labels, labels, all the way!” when facing stiff schemas of what scientists “is” or what artists “is” etc. It’s a hard task by itself to integrate multiple passions and multiple interests into a coherent structure. I don’t need another set of issues because of labels people attach to seemingly creative professions. But limiting myself only to topics consistent with the image of an online scientist became even more frustrating. Therefore expect that this blog (or any other venue I choose to express myself) is going to become a lot more diverse in topics and form.
Posted by Pawel Szczesny on October 28, 2009 in Comments, Research, Science and Art