
Image via Wikipedia
Topic of this post has been sitting in my head for the very long time, but I couldn’t come up with a good enough opening. I’ve found it recently in the comments thread under the post on systems biology by Derek Lowe over at In the Pipeline. Citing Cellbio:
A trick of the human mind has us believe that if we rename something, we have changed the fundamental nature of the beast, but we have not.
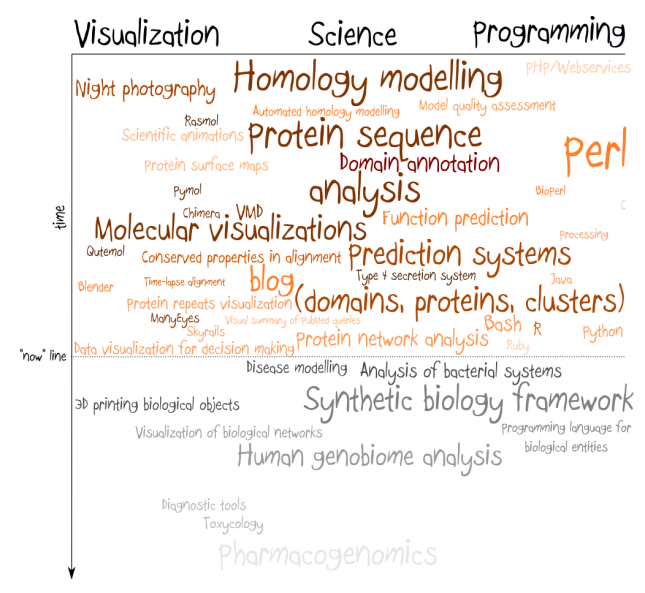
I have taken it out of the context, but it applies very well to current situation in synthetic biology. My enormous frustration with this field comes from the fact that most of so-called synthetic biology is nothing else than genetic engineering with more systematic approach. The whole engineering meme has stuck in people’s head and many of them seem to care more about characterization of the system than about understanding how it works.
If we take a bearing from a car and from a bike, both will differ in shape and very likely one couldn’t be replaced by the other. However, their role and mechanism of work is the same, no matter in which machine we put it (this is BTW what I tried to say in my previous post on BioBricks, but judging from the comments I failed). Mainstream synthetic biology doesn’t seem to be interested in understanding how car and bike works – it’s interested in taking both of them apart as fast as possible, puting labels on the parts and pretend that now we understand how they work. And while this approach can be succesful to a certain extent in engineering, biology, especially synthetic biology, is not engineering, it’s rather a programming.
If we look at the particular component of conserved signalling pathway in two different organisms, its sequence most likely will differ. And for some pairs of organisms sequences of this component stop to be freely exchangable: they need to be mutated to fit particular chassis. Repository of information what works where is a great starting point, but it’s about the time to move further. It’s about the time to express biological systems as sets of functional roles and to build a compiler that transforms an abstract description of biological system into sequence understandable by the particular architecture (organism). This is what I think synthetic biology is all about. It’s designing by understanding.
Formalized language of biological processes sounds like a domain of systems biology, but a compiler certainly doesn’t, so such programming framework could use the best of both worlds. Can you imagine “Hello world” equivalent of a living cell? Or how would you debug program in such language? Sounds like lots of fun.
![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_e.png)








Future of Science on the (ZuiPrezi) map
I’ve just stumbled across map of predictions about global science and innovation created at a recent IFTF workshop in Singapore (the interface is a novel service for online presentation called ZuiPrezi – it looks very promising and I’m waiting for it to come out of private beta). The map contains a few points that resonate with my own scientific interests:
As usuall with such predictions, I feel like many of them are quite conservative – or even schematic. Only very few were completely new to me, but that’s not necessarily a bad thing. It means that actually most of these predictions will turn out to be true in some time. I would like to see something that would immediately blow me away, but on the other hand it’s all relative :).
Related articles by Zemanta
Posted by Pawel Szczesny on September 30, 2008 in Comments, Research
Tags: Energy, Institute for the Future, Microbial fuel cell, Synthetic biology, Technology