
Image via Wikipedia
Topic of this post has been sitting in my head for the very long time, but I couldn’t come up with a good enough opening. I’ve found it recently in the comments thread under the post on systems biology by Derek Lowe over at In the Pipeline. Citing Cellbio:
A trick of the human mind has us believe that if we rename something, we have changed the fundamental nature of the beast, but we have not.
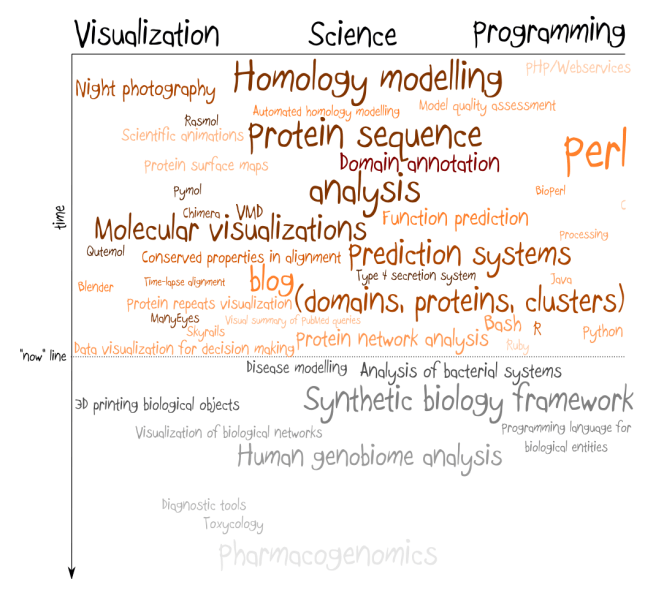
I have taken it out of the context, but it applies very well to current situation in synthetic biology. My enormous frustration with this field comes from the fact that most of so-called synthetic biology is nothing else than genetic engineering with more systematic approach. The whole engineering meme has stuck in people’s head and many of them seem to care more about characterization of the system than about understanding how it works.
If we take a bearing from a car and from a bike, both will differ in shape and very likely one couldn’t be replaced by the other. However, their role and mechanism of work is the same, no matter in which machine we put it (this is BTW what I tried to say in my previous post on BioBricks, but judging from the comments I failed). Mainstream synthetic biology doesn’t seem to be interested in understanding how car and bike works – it’s interested in taking both of them apart as fast as possible, puting labels on the parts and pretend that now we understand how they work. And while this approach can be succesful to a certain extent in engineering, biology, especially synthetic biology, is not engineering, it’s rather a programming.
If we look at the particular component of conserved signalling pathway in two different organisms, its sequence most likely will differ. And for some pairs of organisms sequences of this component stop to be freely exchangable: they need to be mutated to fit particular chassis. Repository of information what works where is a great starting point, but it’s about the time to move further. It’s about the time to express biological systems as sets of functional roles and to build a compiler that transforms an abstract description of biological system into sequence understandable by the particular architecture (organism). This is what I think synthetic biology is all about. It’s designing by understanding.
Formalized language of biological processes sounds like a domain of systems biology, but a compiler certainly doesn’t, so such programming framework could use the best of both worlds. Can you imagine “Hello world” equivalent of a living cell? Or how would you debug program in such language? Sounds like lots of fun.
![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_e.png)













Thinking about RaaS: Research-as-a-Service
Image by Getty Images via Daylife
Instead of disclaimer: this is a bunch of loose thoughts on an element of a possible future of research. I’m only touching some issues here and still I don’t have coherent vision of the commercial side of research. So, feel free to show me I’m very wrong – you’ll save me lots of time of coming to your conclusions :).
According to Wikipedia, Everything as a Service is a:
While the most common example is SaaS – Software as a Service, this concept can be applied to other functions such as communication, infrastructure or data (the last one sounds very interesting). It recently occurred to me, that investments (or maybe I should call these partnerships) of biotech and pharma companies in academic research institutions are good examples of RaaS, Research as a Service. I think every situation where the research is done after the agreement (buying or licensing patented innovation doesn’t qualify) can be called RaaS.
Why
To a company, there are obvious advantages of hiring scientists to get the research done, but I think there also would be plenty of good sides of such arrangement for us (of course I have no experience yet). Probably the biggest plus would be money and ability to get them in somehow predictable manner. I think it’s also important to be stretched intellectually from time to time (I assume that easy things aren’t worth outsourcing).
Many flavors of RaaS
Paying to an academic institution to come up with a new drug candidate is only one of many types of RaaS. There’s researching a given problem (something Innocentive or Nine Sigma are coordinating), coming up with an innovation (drug candidate example), providing expertise (consulting) or innovating and delivering (designing, building and implementing new machine, workflow or pipeline). We could find examples of all types happening everyday, but probably not in all scientific fields. Delivering something in biology is usually quite expensive and time consuming, while consulting gigs in quantum physics don’t appear all that often.
The point is that all these RaaS flavors can and are applied to academic institutions. In other words, many researchers provide commercial services using time and equipment paid from taxpayers money. And I think it’s not an issue – even more: it should be finally admitted and accepted (so we could get rid of the artificial division of researches into academic and all others; but that’s another story), and organized, so we could provide such services easier and more often.
Resources all over the place
The Health Commons project aims at building a framework that could help in sharing and organising research process aiming at developing new drugs. We seem to have lots of elements of such environment in place – we have many (or even too many 😉 ) scientists, some service providers, data centers and some work done on standards of operations and information exchange. If we forget about drug development, not much actually changes. We have workforce, some services aimed at researchers and lots of tools that help in communication in both directions.
Here’s example of research scenario: if I were to market a genetic test that identifies mutations resulting in oversensitivity or resistance to a drug (something which I believe will be the next hit after screening for disease markers), the whole research part wouldn’t require any significant involvement from my side. CRO (contract research organization) would take care of identifying patients with specific conditions, sequencing company would get me their genomes (currently $5000 each, but the price is dropping very fast) and as far as I know bioinformatics community, finding people to analyze the data wouldn’t be an issue at all. While such scenario is a bit too optimistic (I skipped lawyers in the process), we already have resources to make it happen.
Where is it going?
I imagine future RaaS provider as a small company (I’m not yet sure if a non-profit organization is a better fit for people interested in doing research; also, I don’t know how fast the issue of academic-commercial blur can be solved) made by a few scientists from different but closely related fields. The reason I see it small, is about mobility. And I don’t mean here physical mobility (which BTW may be required on some occasions) but mobility of focus – the main advantage of small organizations.
I imagine such company would be able to do consulting (and data analysis, maybe on the RedMonk model) and innovate at a software level. It would be able to do the work on site (small group again) and deliver the results quick (“bursty work”).
Pieces of this vision come from old Deepak’s posts and many FriendFeed discussions. I actually think about putting it into practice. What do you think I am missing here (other than marketing 😉 )?
Posted by Pawel Szczesny on November 3, 2008 in Comments, Research
Tags: Business, Cloud computing, Intellectual property, Knowledge Management, raas, Research, Saas, service, Software as a service, Venture capital