Images of molecules
Published or almost published images
OmpA-like protein from P. gingivalis
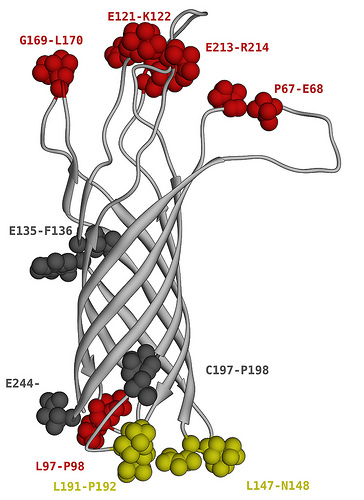
Membrane anchor part of trimeric autotransporter adhesins…
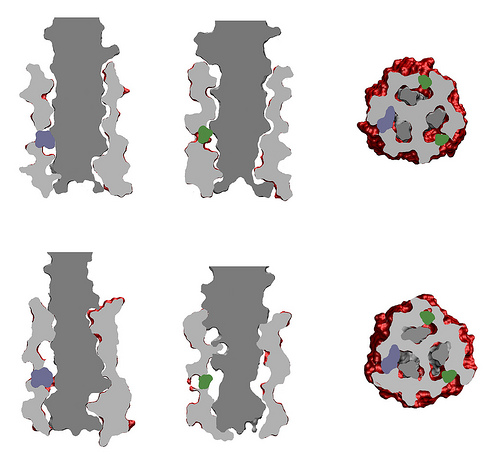
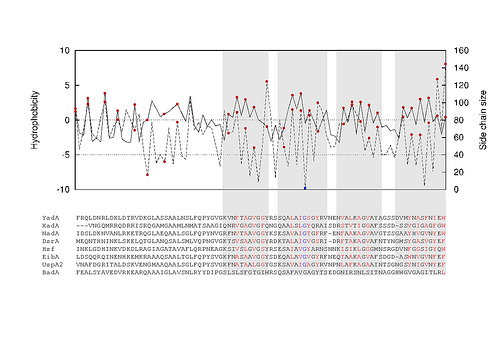
… and its alignment
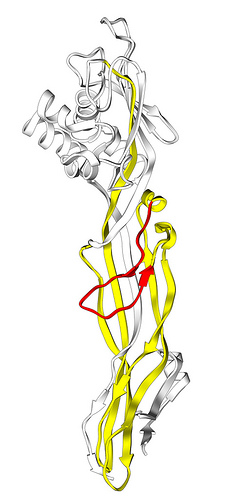
Aerolysin-like protein
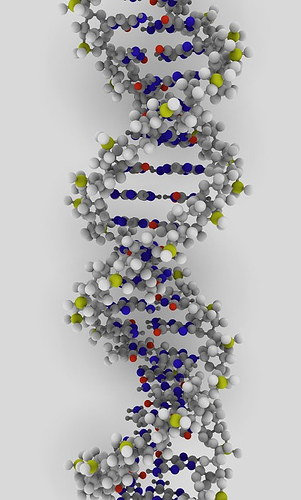
DNA
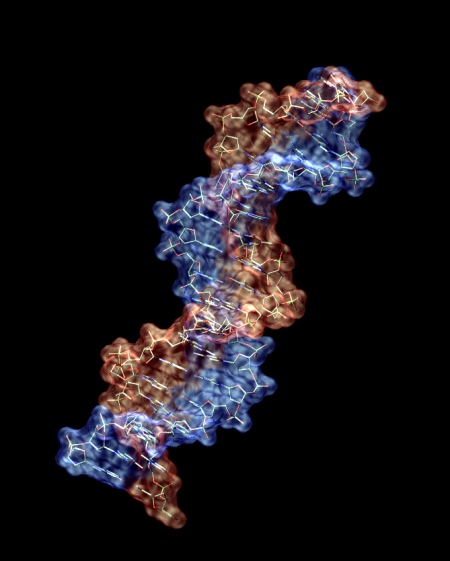
More artistic representations
B-DNA:

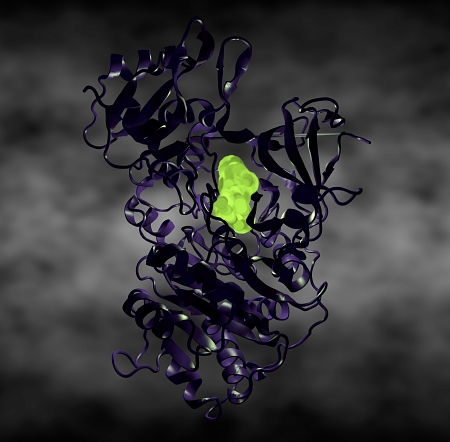
Luciferase:
Hemoglobin:
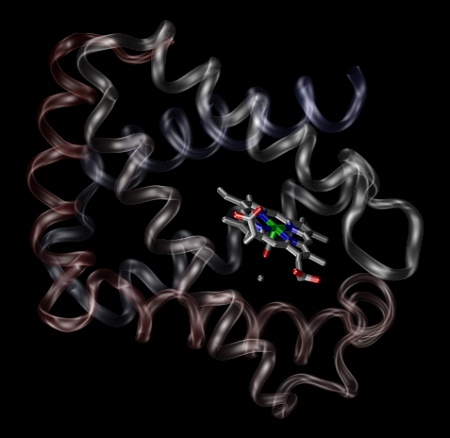

Molecular manipulator designed by Dr. Drexler from imm.org.
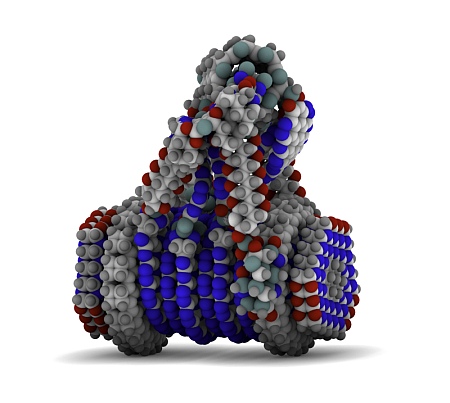
Neisserial surface protein A (NspA)

Importing PDB file into Blender
Because it seems some of the readers may be interested in importing PDB file into Blender, here’s a short guide how to do it. Actually, there are two approaches to do it. First is represented by image of molecular manipulator, the second by image of NspA (two last pictures above).
Use pdb2blend script:
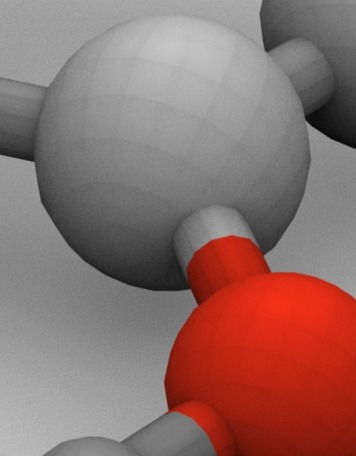
Download pdb2blend converter, open it in Blender and run it (Alt+P). Now, in the script window you can choose some options, like representation (balls, sticks and balls or sticks) or choose level of refinement. After import, the structure in chosen representation will appear in the 3D window with atoms colored according to schema in the script. There’s actually one caveat – spheres prepared with this script when rendered with ambient occlusion exhibit a weird pattern seen below. I’m not Blender expert so I have no idea how to solve it.
Second way of importing PDB structure into Blender in almost any representation is to use STL format. At least two molecular viewers are able to export structure in STL format: VMD and PMV (Python Molecular Viewer). I use PMV more often because it can export STL file with custom quality. Unfortunately, colors are not preserved in STL files from both programs (or Blender cannot read it, I’m not really sure). So, the you open PDB file in either of this programs, choose representation and then export/save as STL (in VMD this option is available under “Render”). After import the only thing left is to remove double vertices (edit mode, select all, edit -> vertices -> remove doubles). Voilla. You have molecular structures inside Blender.






Deepak
October 28, 2007 at 18:52
Where did you do the surface renderings? PyMol?
freesci
October 28, 2007 at 19:29
First image, right? In VMD, using one of “glass” materials.
Marco
November 9, 2007 at 16:33
Luciferase is simply awesome! 🙂
freesci
November 10, 2007 at 08:43
Thanks, Marco.
Thomas Scoville
March 13, 2008 at 02:39
Groovy renderings. Cool.
Pawel Szczesny
March 16, 2008 at 17:24
Thanks, Thomas.
ed
March 25, 2008 at 20:15
how did you make “Neisserial surface protein A (NspA)”?
Pawel Szczesny
March 25, 2008 at 20:41
Hi Ed, in Blender, with ambient occlusion switched on.
Lenin
March 29, 2008 at 07:02
Nice images!
The last two are my favorites!
renee albrecht
November 8, 2008 at 16:51
perfect
Daymon Balser
January 16, 2009 at 10:45
I’ll have to say that this is art in it’s truest form. A Quantum Celebration of Life.
nah sivar
February 14, 2009 at 17:25
Could you provide pointers (or tutorials) to get one started with playing with PDB in Blender and creating images like the one above ! (they are awesome )
it will be nice if the importing of pdb, & doing things with it is presented for someone really new to Blender.
akin to starting with Blender by playing with PDB strcutures ?!
someone plz.,
nah sivar
February 14, 2009 at 17:29
could you kindly elaborate on the steps involved in creating such beautiful images ?
from sample PDB to a cool picture – steps or sample protocol will be really helpful.
it will also have many people started with Blender. I am willing to put in time and write some a document if you will help me in getting started with it (i have played with Pymol, Photoshop etc., but new to 3D-rendering and just now installed Blender (under Vista )
the pdb2blend conversion script had some deprecation warning in the new Blender (current version)
if you know of other developments/ or programs to be used for such rendering it will be great if you can share them as well
Malte Reimold
March 3, 2009 at 12:59
The pdb2blend script works fine with Blender 2.48.
Some problems arise from pdb-files which do not comply with the pdb file format conventions. I did not introduce any error handling for this case so the script may be not functional for some pdb-files.
This ambient occlusion problem is new to me and I was not able to reproduce it. It looks like the mesh would have been set to solid.
Farid
October 2, 2009 at 18:15
Dear Pawel,
I am looking for a company logo and I like the structure of your Neisserial surface protein A (NspA) ut I would like to highlight one residue(ball and stick which glows geen) and the words ‘HEPA technology’ below it.
I don’t mind paying a fee for your service….
best wishes,
Farid
Pella86
November 8, 2011 at 13:13
Hello, I’m not an expert of blender, but “There’s actually one caveat – spheres prepared with this script when rendered with ambient occlusion exhibit a weird pattern seen below”, this can simply be corrected if you select the mesh-> edit mode-> set smooth. This would actually take away the solid pattern of the sphere, or you could add a subsurface, but then the edges between the atoms and the bonds will not be sharp.
hope that helps
Pella
top mistakes
December 11, 2011 at 11:03
Your site is really cool to me and your topics are very relevant. I was browsing around and came across something you might find interesting. I was guilty of 3 of them with my sites. “99% of website owners are committing these 5 errors”. http://bit.ly/tBfFqz You will be suprised how fast they are to fix.
94Bob
August 9, 2017 at 17:46
Hello admin, i must say you have high quality posts here.
Your website should go viral. You need initial traffic only.
How to get it? Search for; Mertiso’s tips go viral